10 Atp5a Qpcr Primer Tips For Accurate Results

The ATP5A gene, encoding a subunit of the mitochondrial ATP synthase, plays a crucial role in energy production within cells. Quantitative Polymerase Chain Reaction (qPCR) is a sensitive technique used to measure the expression levels of this gene. To ensure accurate results in ATP5A qPCR experiments, careful primer design and optimization are essential. Here, we provide 10 expert tips for designing and using qPCR primers for the ATP5A gene, focusing on specificity, efficiency, and reliability.
Understanding the Basics of qPCR Primer Design
Before diving into the tips, it’s essential to understand the fundamentals of qPCR primer design. Specificity is key to avoiding non-specific binding and ensuring that the primers only amplify the target sequence. Primer melting temperature ™ should be between 58°C and 60°C for optimal annealing. The primer length typically ranges from 18 to 24 nucleotides, and GC content should be around 50% for efficient amplification.
Tips for Optimizing ATP5A qPCR Primers
1. Use a reliable primer design tool: Utilize software like Primer3 or NCBI Primer-BLAST to design primers that meet the optimal criteria for Tm, length, and GC content. These tools help in avoiding repetitive sequences and ensuring specificity to the ATP5A gene.
2. Check for secondary structures: Use tools like mfold to predict potential secondary structures in the primers. Secondary structures can hinder primer binding and affect qPCR efficiency.
3. Optimize primer concentration: The optimal primer concentration can vary. Typically, a range of 0.1 to 0.5 μM is recommended. Higher concentrations can lead to non-specific binding and primer-dimer formation.
4. Choose the right qPCR mix: Select a qPCR mix that is compatible with your primer design and experimental conditions. Some mixes are optimized for specific dyes or instruments.
5. Validate primer specificity: Perform a melting curve analysis after qPCR to ensure that the primers are specific to the ATP5A gene. A single peak indicates specificity, while multiple peaks suggest non-specific binding.
| Primer Characteristic | Optimal Value |
|---|---|
| Length | 18-24 nucleotides |
| GC Content | Around 50% |
| Tm | 58°C - 60°C |
| Concentration | 0.1 - 0.5 μM |

Advanced Considerations for ATP5A qPCR
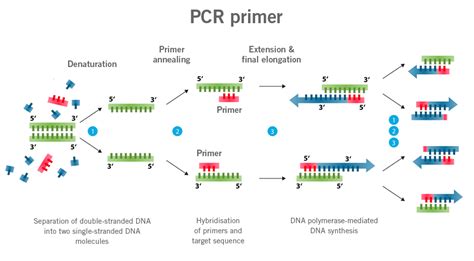
6. Use reference genes for normalization: Normalize ATP5A expression data with reference genes to account for variations in cDNA input and PCR efficiency. Common reference genes include ACTB, GAPDH, and PPIA.
7. Optimize the annealing temperature: While the default annealing temperature is often 60°C, optimizing this temperature for your specific primers can improve specificity and efficiency.
8. Consider using probe-based qPCR: For increased specificity, especially when dealing with genes that have pseudogenes or highly similar sequences, consider using probe-based qPCR assays like TaqMan.
9. Run replicate samples: Include technical replicates (multiple qPCR reactions from the same cDNA sample) and biological replicates (samples from different subjects or conditions) to ensure reliable and reproducible results.
10. Analyze data with appropriate software: Use software like ABI Prism or BioRad CFX Maestro to analyze qPCR data. These programs can help in determining amplification efficiency, calculating expression levels, and performing statistical analysis.
What are the most critical factors in designing qPCR primers for the ATP5A gene?
+The most critical factors include specificity, primer melting temperature (Tm), primer length, and GC content. Ensuring these factors are optimized helps in achieving efficient and specific amplification of the target sequence.
How can I validate the specificity of my qPCR primers for ATP5A?
+Perform a melting curve analysis after the qPCR reaction. A single peak in the melting curve indicates that the primers are specific to the ATP5A gene. Additionally, sequencing the PCR product or running it on a gel can further confirm specificity.
In conclusion, designing and optimizing qPCR primers for the ATP5A gene requires careful consideration of several factors, including primer specificity, Tm, length, GC content, and concentration. By following these 10 expert tips and considering advanced aspects of qPCR such as reference genes, annealing temperature, and replicate samples, researchers can ensure accurate and reliable results in their ATP5A qPCR experiments.