12 Peromyscus Dna Polymerase Hacks For Faster Results

The Peromyscus genus, commonly known as deer mice, has been a subject of interest in molecular biology due to its unique genetic characteristics. One area of research involves the DNA polymerase enzymes derived from these organisms, which have shown potential in enhancing PCR (Polymerase Chain Reaction) efficiency and fidelity. DNA polymerases are essential for DNA replication and repair, and their properties can significantly impact the outcome of molecular biology experiments. Here, we will explore 12 hacks for utilizing Peromyscus DNA polymerase to achieve faster results in PCR reactions.
Understanding Peromyscus DNA Polymerase

Peromyscus DNA polymerase, like other DNA polymerases, is responsible for adding nucleotides to a growing DNA chain during replication. Its high processivity and fidelity make it an attractive choice for PCR applications. Processivity refers to the ability of the polymerase to synthesize long DNA strands without dissociating from the template, while fidelity is its accuracy in incorporating the correct nucleotides. The unique properties of Peromyscus DNA polymerase, including its thermostability, allow it to withstand the high temperatures required for PCR, making it a valuable tool in molecular biology research.
Optimizing Reaction Conditions
To achieve faster results with Peromyscus DNA polymerase, optimizing the reaction conditions is crucial. This includes adjusting the primer concentration, template DNA amount, and enzyme concentration. A balanced reaction mixture ensures that the polymerase can efficiently bind to the template and synthesize the new DNA strand. Moreover, the choice of buffer components and ions can affect the enzyme’s activity and stability. For instance, the presence of magnesium ions (Mg2+) is essential for the polymerase activity, but excessive amounts can inhibit the reaction.
| Reaction Component | Optimal Condition |
|---|---|
| Primer Concentration | 0.1-1.0 μM |
| Template DNA Amount | 10-100 ng |
| Enzyme Concentration | 0.5-2.0 units/μL |
| Magnesium Ion Concentration | 1.5-2.5 mM |
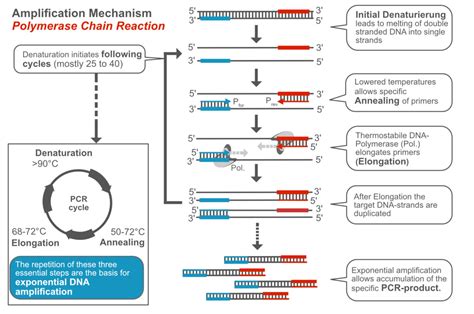
Enhancing Reaction Efficiency

Besides optimizing reaction conditions, several strategies can be employed to enhance the efficiency of PCR reactions using Peromyscus DNA polymerase. PCR enhancers, such as betaine or dimethyl sulfoxide (DMSO), can improve the reaction by stabilizing the DNA template, reducing secondary structure formation, and increasing the melting temperature of the primers. Additionally, annealing temperature optimization is critical for specificity, as it affects the binding of primers to the template DNA. A touchdown PCR protocol, where the annealing temperature is gradually decreased, can help in identifying the optimal annealing temperature for the primers.
Minimizing Inhibitors and Contaminants
The presence of inhibitors or contaminants in the reaction mixture can significantly hinder the PCR efficiency. Template DNA purification methods, such as silica column-based purification or phenol-chloroform extraction, can help remove contaminants. Moreover, the use of inhibitor-resistant DNA polymerases or additives like Tween 20 can mitigate the effects of residual inhibitors. It is also essential to handle PCR reactions in a PCR clean environment to prevent cross-contamination with exogenous DNA.
- Purify template DNA using silica column-based methods.
- Use inhibitor-resistant DNA polymerases or additives.
- Handle reactions in a PCR clean environment.
What are the key factors to consider when optimizing PCR reactions with Peromyscus DNA polymerase?
+The key factors include primer concentration, template DNA amount, enzyme concentration, magnesium ion concentration, and annealing temperature. Additionally, the use of PCR enhancers, hot-start PCR, and minimizing inhibitors and contaminants can significantly impact the reaction efficiency and specificity.
How can the processivity and fidelity of Peromyscus DNA polymerase be advantageous in PCR applications?
+The high processivity of Peromyscus DNA polymerase allows for the efficient synthesis of long DNA strands, reducing the need for multiple primer binding events and thus increasing the reaction speed. Its high fidelity ensures that the synthesized DNA strands have minimal errors, which is crucial for applications where accuracy is paramount, such as in diagnostic PCR and cloning.
In conclusion, Peromyscus DNA polymerase offers several advantages in PCR applications due to its unique properties. By optimizing reaction conditions, enhancing reaction efficiency, and minimizing inhibitors and contaminants, researchers can achieve faster and more accurate results. Understanding the specifics of how to utilize Peromyscus DNA polymerase effectively can significantly improve the outcomes of molecular biology experiments, contributing to advancements in fields such as genetics, biotechnology, and diagnostics.