Mismatch Repair Guide: Dna Correction Mastery
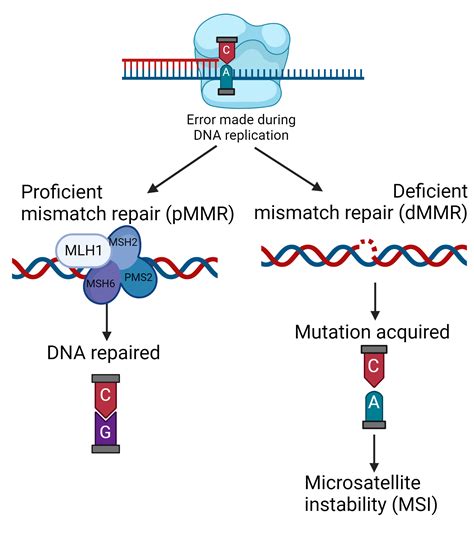
The mismatch repair (MMR) system is a vital component of the DNA repair machinery, responsible for correcting errors in DNA replication and recombination. This complex process involves the recognition and correction of mismatched bases, insertions, and deletions, ensuring the maintenance of genome stability. The MMR system is conserved across prokaryotes and eukaryotes, with a high degree of similarity in the core components and mechanisms. In this comprehensive guide, we will delve into the intricacies of the MMR system, exploring its key components, mechanisms, and implications for genome stability and disease.
Introduction to Mismatch Repair

The MMR system was first discovered in Escherichia coli in the 1970s, and since then, it has been extensively studied in various organisms. The core components of the MMR system include the MutS and MutL proteins, which are responsible for recognizing and processing mismatched DNA. The MutS protein recognizes mismatched bases and forms a complex with the MutL protein, which then recruits other proteins to correct the error. The MMR system is essential for maintaining genome stability, as defects in this pathway can lead to increased mutation rates, genomic instability, and cancer.
Key Components of the MMR System
The MMR system involves a complex interplay of proteins, including:
- MutS: Recognizes mismatched bases and forms a complex with MutL
- MutL: Recruits other proteins to correct the error
- MutH: An endonuclease that cleaves the mismatched strand
- Helicase: Unwinds the DNA double helix
- Exonuclease: Degrades the mismatched strand
- DNA polymerase: Synthesizes new DNA
- DNA ligase: Seals the nick in the DNA backbone
| Component | Function |
|---|---|
| MutS | Recognizes mismatched bases |
| MutL | Recruits other proteins to correct the error |
| MutH | Cleaves the mismatched strand |
Mechanisms of Mismatch Repair
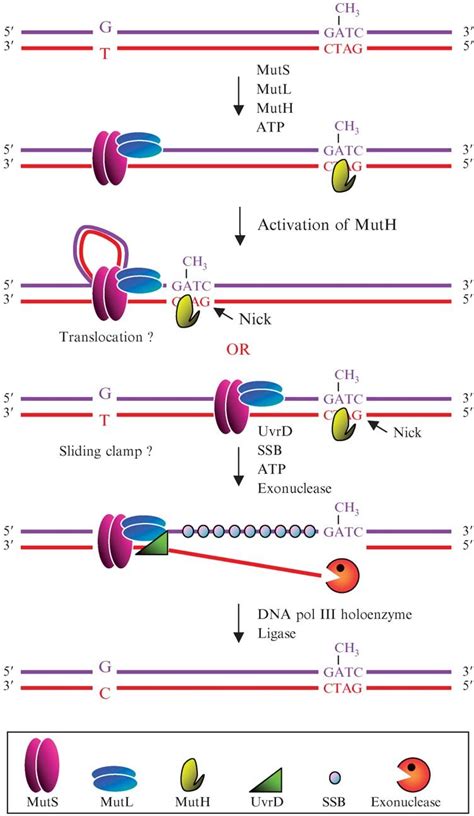
The MMR system involves a complex series of steps, including:
- Recognition of mismatched bases by MutS
- Formation of a complex with MutL
- Recruitment of other proteins to correct the error
- Cleavage of the mismatched strand by MutH
- Unwinding of the DNA double helix by helicase
- Degradation of the mismatched strand by exonuclease
- Synthesis of new DNA by DNA polymerase
- Sealing of the nick in the DNA backbone by DNA ligase
The MMR system is highly efficient, with a repair efficiency of up to 90%. However, defects in the MMR system can lead to increased mutation rates, genomic instability, and cancer.
Implications of Mismatch Repair Defects
Defects in the MMR system have been implicated in various diseases, including:
- Hereditary nonpolyposis colorectal cancer (HNPCC)
- Lynch syndrome
- Turcot syndrome
- Li-Fraumeni syndrome
| Disease | MMR Defect |
|---|---|
| HNPCC | Defects in MLH1 or MSH2 |
| Lynch syndrome | Defects in MLH1, MSH2, MSH6, or PMS2 |
| Turcot syndrome | Defects in MLH1 or MSH2 |
Future Directions and Implications

The MMR system is a critical component of the DNA repair machinery, and understanding its intricacies can provide valuable insights into the maintenance of genome stability and the development of diseases. Future research should focus on:
- Elucidating the molecular mechanisms of the MMR system
- Understanding the implications of MMR defects in disease
- Developing therapeutic strategies to target MMR-deficient cancers
What is the role of the MMR system in maintaining genome stability?
+The MMR system is responsible for correcting errors in DNA replication and recombination, ensuring the maintenance of genome stability. Defects in the MMR system can lead to increased mutation rates, genomic instability, and cancer.
What are the implications of MMR defects in disease?
+Defects in the MMR system have been implicated in various diseases, including HNPCC, Lynch syndrome, Turcot syndrome, and Li-Fraumeni syndrome. Understanding the implications of MMR defects can provide valuable insights into the development of diseases and the importance of maintaining genome stability.